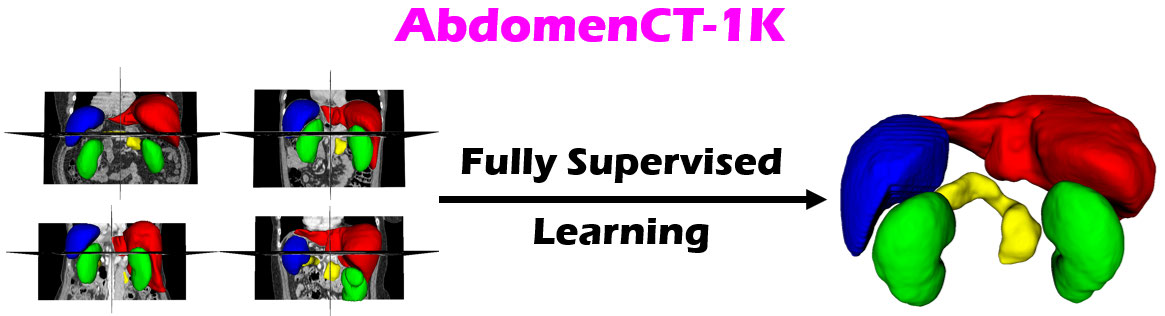
Dataset Introduction
The dataset is adapted from existing benchmark datasets [1-6] by annotating four organs in each case, including liver (label=1), kidney (label=2), spleen (label=3), and pancreas (label=4).
More details are in the following paper:
@ARTICLE{abdomenct-1k,
author={Ma, Jun and Zhang, Yao and Gu, Song and Zhu, Cheng and Ge, Cheng and Zhang, Yichi and An, Xingle and Wang, Congcong and Wang, Qiyuan and Liu, Xin and Cao, Shucheng and Zhang, Qi and Liu, Shangqing and Wang, Yunpeng and Li, Yuhui and He, Jian and Yang, Xiaoping},
journal={IEEE Transactions on Pattern Analysis and Machine Intelligence},
title={AbdomenCT-1K: Is Abdominal Organ Segmentation a Solved Problem?},
year={2022},
volume={44},
number={10},
pages={6695-6714},
doi={10.1109/TPAMI.2021.3100536}
}Download
- Zenodo
- Baidu Net Disk (PW: 2021)
For fair comparison of different methods, we also hold a hidden testing set (50 cases) from a new medical center. Please package your algorithm as a Docker and submit it to us. The submitted Docker should be named TeamName.tar.gz and we will evaluate it with the following two commands:
docker image load < teamname.tar.gz
docker container run --gpus "device=1" --name teamname --rm \
-v $PWD/inputs/:/workspace/inputs/ \
-v $PWD/teamname_outputs/:/workspace/outputs/ \
teamname:latest /bin/bash -c "sh predict.sh"
Reference
[1] N. Heller, F. Isensee, K. H. Maier-Hein, X. Hou, C. Xie, F. Li, Y. Nan, G. Mu, Z. Lin, M. Han et al., “The state of the art in kidney and kidney tumor segmentation in contrast-enhanced ct imaging: Results of the kits19 challenge,” Medical Image Analysis, vol. 67, p. 101821, 2021.
[2] A. L. Simpson, M. Antonelli, S. Bakas, M. Bilello, K. Farahani, B. Van Ginneken, A. Kopp-Schneider, B. A. Landman, G. Litjens, B. Menze et al., “A large annotated medical image dataset for the development and evaluation of segmentation algorithms,” arXiv preprint arXiv:1902.09063, 2019.
[3] K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips, D. Maffitt, M. Pringle et al., “The cancer imaging archive (tcia): maintaining and operating a public information repository,” Journal of Digital Imaging, vol. 26, no. 6, pp. 1045–1057, 2013.
[4] P. Bilic, P. F. Christ, E. Vorontsov, G. Chlebus, H. Chen, Q. Dou, C.-W. Fu, X. Han, P.-A. Heng, J. Hesser et al., "The liver tumor segmentation benchmark (lits)," arXiv preprint arXiv:1901.04056, 2019.
[5] H. R. Roth, A. Farag, E. B. Turkbey, L. Lu, J. Liu, and R. M. Summers, “Data from pancreas-CT,” The Cancer Imaging Archive, 2016.
[6] H. R. Roth, L. Lu, A. Farag, H.-C. Shin, J. Liu, E. B. Turkbey, and R. M. Summers, “Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation,” in International Conference on Medical Image Computing and Computer-assisted Intervention, 2015, pp. 556–564.